How many of you that are working with artificial DNA sets and have to use the tools that are available over the Web have to edit the output because the FASTA header of such programs does not correspond to your needs, the output is not in FASTA format or the lazy reason to copy and paste the output sequences inside a manually generated file because it depends on important ATP molecules that can be used in another tasks.
STEP 3 <- Please type the number of iterations (How many random sequences do you want)
In this Tutorial I want to generate 100 artificial sequences.
STEP 4 <- Please type the length of the random DNA strings (how many nucleotides length)
In this Tutorial I want each sequence to be 100 nucleotides long
STEP 6 <- Please, type the name of the fasta header for each sequence (is not necessary to put the >)
In this Tutorial, I want that the FASTA header be: "just a bunch of random sequences of equal random prob nt distribution"
STEP 7 <- Please, type the name of the output file:
In this Tutorial, I want to name it "ran_set_1.fa"
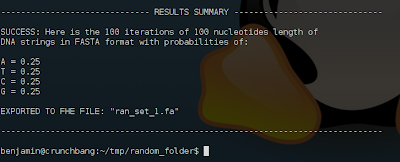
STEP 8 <- Enjoy the results
Another example:
Now I want to generate 100 random DNA sequences of 100 nucleotides length with a nucleotide distribution of "A=0.7, T=0.2, G=0.1 and C=0.0" with the FASTA header of "random dna sequence with too many As" in a file called "random_set_A_rich.fa".
Now, take a look of the output folder:
Do you see it :D, now we got two output files in FASTA format of random made sequences that follows a probability distribution of nucleotides given by us.
NOTE: The yellow highlighting just mean the differences of the content of A nucleotides among the two files showing us that the probability distribution obviously has an impact on the nucleotide composition of the random generated DNA strings.
Benjamin
Well, this Perl script called "random_dna_strings.pl" can be downloaded by clicking the link.
This script takes arguments given by the user such the nucleotide frequencies of each nucleotide (in a scale from 0.0 to 1.0), generates a "n" number of sequences of "n" length with a FASTA header also given by the user and finally prints an output file in FASTA format.
So, here is a short Tutorial about how to use it:
Well, imagine that we want to generate 100 random DNA sequences of 100 nucleotides length with a nucleotide distribution equal to every nucleotide, this is "A=0.25, T=0.25, G=0.25 and C=0.25" with the FASTA header of "just a bunch of random sequences of equal random prob nt distribution" in a file called "ran_set_1.fa".
STEP 1 <- Open a Terminal inside the output folder (where you want to put the output file)
STEP 2 <- Execute the Perl script (This time, I copied the script to my bin folder, if you have questions about how to run your scripts this way, please visit this entry.
Command:
$ random_dna_strings.plIn this Tutorial I want to generate 100 artificial sequences.
In this Tutorial I want each sequence to be 100 nucleotides long
STEP 5 <- Please type the probability distribution of A, T, G and C content:
In this Tutorial I want each nucleotide got an equal probability distribution (this means 1/4 of probability of A,T,G or C at each string position)
In this Tutorial I want each nucleotide got an equal probability distribution (this means 1/4 of probability of A,T,G or C at each string position)
In this Tutorial, I want that the FASTA header be: "just a bunch of random sequences of equal random prob nt distribution"
STEP 7 <- Please, type the name of the output file:
In this Tutorial, I want to name it "ran_set_1.fa"
Now I want to generate 100 random DNA sequences of 100 nucleotides length with a nucleotide distribution of "A=0.7, T=0.2, G=0.1 and C=0.0" with the FASTA header of "random dna sequence with too many As" in a file called "random_set_A_rich.fa".
NOTE: The yellow highlighting just mean the differences of the content of A nucleotides among the two files showing us that the probability distribution obviously has an impact on the nucleotide composition of the random generated DNA strings.
Benjamin









No comments:
Post a Comment
Note: Only a member of this blog may post a comment.